Erica Forsberg
Assistant Professor, Analytical, Bioanalytical Chemistry
office: CSL 401
phone: 619-594-5806
email: eforsberg@sdsu.edu

Forsberg Group Page
Curriculum Vitae
- Assistant Professor, San Diego State University, 2017-present
- Post-doctoral Fellow, The Scripps Research Institute, 2015-2017
- Ph.D. Chemical Biology, McMaster University, 2015
- B.Sc. Chemistry, summa cum laude, McMaster University, 2008
Research Interests
Have you ever had a gut feeling? Well, there may be a scientific explanation to this phenomenon! Within the last decade, there has been a significant paradigm shift in our understanding of the human microbiome and its impact on human health. Microbial cells outnumber human cells 10 to 1, and we have only recently begun to understand their impact on producing metabolites that affect us. One uncharacterized interaction is the interface between microbial metabolites and neurological function, or the gut-brain axis. Gut metabolites, such as serotonin, impact our feelings of hunger and satiety, but can play a major role in diseases such as irritable bowel syndrome, obesity, and colon cancer, and even anxiety and depression.Culturing Anaerobic Microorganisms in a Human Gut Model
The Forsberg lab looks to decipher biological mechanisms of gut microbiota using analytical techniques, particularly mass spectrometry (MS) based metabolomics. A key to understanding mechanistic perturbations in a biological system requires growing anaerobic bacteria in a system closely resembling the human gut. Anaerobic culturing techniques facilitate growth studies looking at the effects that diet, antibiotics and xenobiotics have on microbial growth, metabolism and signaling. Culturing gut microorganisms is challenging. Mimicking conditions inside the human intestinal tract has yet to be perfected in the laboratory setting, and metabolite extraction methods need further development.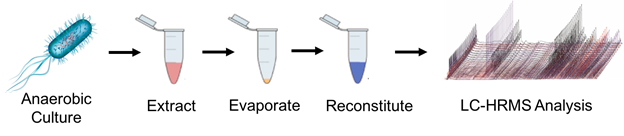
Figure 1. Gut microbe culturing and analysis workflow requires optimization of anaerobic conditions, the addition of a stress condition, such as antibiotics, for comparison, metabolite extraction followed by liquid chromatography high resolution mass spectrometry (LC-HRMS).
Metabolomics and Bioinformatics
Metabolomics focuses on bioactive small molecules, such as central carbon metabolites involved in biosynthetic pathways. When a well-characterized biological system is stressed, metabolites may become dysregulated. We can analyze these alterations in metabolic pathways via tandem mass spectrometry, which fragments analytes and tracks specific product ions. Quantifying metabolite changes can illuminate alterations in protein activity stemming from inhibition, activation, or changes in regulation. When the stress effects are unknown or the pathways are uncharacterized, microbial metabolism is far more complicated and requires global or untargeted metabolomics, which profiles the entire metabolome in an unbiased manner. However, the complexity of samples results in tens of thousands of detected metabolites and requires intense data analysis using bioinformatics. Although there are some excellent platforms available for analyzing untargeted data (e.g. MetaboScape, XCMS Online, MetaboAnalyst), they have their limits. There is still a need to improve data integration and statistical methods from multiple organisms. Our goal is to effectively understand interactions between multiple microbial sources, or microbiota and human host, in a single streamlined analysis.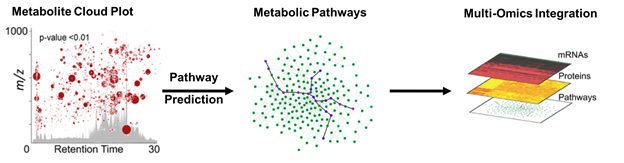
Figure 2. Example of bioinformatic analysis workflow for large untargeted metabolomic datasets in conjunction with multi-omics data using XCMS Online. These powerful tools will help us build a system to analyze multiple microorganisms and the human host simultaneously.
Separation and Identification of Complex Biological Mixtures
High resolution (HR) instruments such as quadrupole time of flight (QTOF) or Orbitrap mass spectrometers accurately and precisely measure a large dynamic range of molecular weight analytes, from small molecules to large proteins. Our lab utilizes HR-MS coupled to liquid chromatography (LC) to identify compounds in complex biological mixtures. LC provides a means to separate a biological sample to achieve high metabolomic coverage over both hydrophobic and polar compounds. Although LC powerfully separates a large range of analytes without modifying the sample, it is less reproducible and slower than gas chromatography (GC). We will develop novel methods employing a wide variety of stationary phases and sample introduction techniques to maximize efficiency and reproducibility of separations at low cost.Selected publications
- "Towards predicting gut microbial metabolism: Integration of flux balance analysis and untargeted metabolomics,"
Kuang, E; Marney, M.; Cuevas, D.; Edwards, R.A.; Forsberg, E.M.,
Metabolites 10, 156 (2020). (10.3390/metabo10040156.) - "A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome,"
Riva, A.; Kuzyk, O.; Forsberg, E.; Siuzdak, G.; Pfann, C.; Herbold, C.; Daims, H.; Loy, A.; Warth, B.; Berry, D.,
Nature Communications 10, 4366 (2019). (10.1038/s41467-019-12413-0.) - "Fluorinated Gold Nanoparticles for Nanostructure Imaging Mass Spectrometry,"
Amelia Palermo, Erica M. Forsberg, Benedikt Warth, Aries E. Aisporna, Elizabeth Billings, Ellen Kuang, H. Paul Benton, David Berry, and Gary Siuzdak,
ACS Nano 12, 6938-6948 (2018). (10.1021/acsnano.8b02376.) - "Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online,"
Forsberg, E.; Huan, T.; Rinehart, D.; Benton, H. P.; Hilmers, B.; Warth, B.; Siuzdak, G.,
Nature Protocols 13, 633-651 (2018). (doi: 10.1038/nprot.2017.151.) - "Systems biology guided by XCMS Online metabolomics,"
Huan, T.; Forsberg, E. M.; Rinehart, D.; Johnson, C. H.; Ivanisevic, J.; Benton, H. P.; Fang, M.; Aisporna, A.; Hilmers, B.; Poole, F. L.; Thorgersen, M. P.; Adams, M. W. W.; Krantz, G.; Fields, M. W.; Robbins, P. D.; Niedernhofer, L. J.; Ideker, T.; Majumder, E. L.; Wall, J. D.; Rattray, N. J. W.; Goodacre, R.; Lairson, L. L.; Siuzdak, G.,
Nat. Meth. 14, 461-462 (2017). (doi: 10.1038/nmeth.4260..) - "Evaluation of the safety and immunomodulatory effects of sargramostim in a randomized, double-blind phase 1 clinical Parkinson's disease trial,"
Gendelman, H. E.; Zhang, Y.; Santamaria, P.; Olson, K. E.; Schutt, C. R.; Bhatti, D.; Shetty, B. L. D.; Lu, Y.; Estes, K. A.; Standaert, D. G.; Heinrichs-Graham, E.; Larson, L.; Meza, J. L.; Follett, M.; Forsberg, E.; Siuzdak, G.; Wilson, T. W.; Peterson, C.; Mosley, R. L.,
npj Parkinson's Disease 3, 10 (2017). (doi: 10.1038/s41531-017-0013-5..) - "Staying Alive: Measuring Intact Viable Microbes with Electrospray Ionization Mass Spectrometry,"
Forsberg, E.; Fang, M.; Siuzdak, G.,
Journal of The American Society for Mass Spectrometry 28, 14-20 (2017). (doi: 10.1007/s13361-016-1440-y..) - "Global Isotope Metabolomics Reveals Adaptive Strategies for Nitrogen Assimilation.,"
Kurczy, M. E.; Forsberg, E. M.; Thorgersen, M. P.; Poole, F. L.; Benton, H. P.; Ivanisevic, J.; Tran, M. L.; Wall, J. D.; Elias, D. A.; Adams, M. W. W.; Siuzdak, G.,
ACS Chemical Biology 11, 1677-1685 (2016). (doi: 10.1021/acschembio.6b00082..) - "Solid-Phase Biological Assays for Drug Discovery,"
Forsberg, E. M.; Sicard, C.; Brennan, J. D.,
Annual Review of Analytical Chemistry 7, 337-359 (2014). (doi: 10.1146/annurev-anchem-071213-020241..) - "Bio-Solid-Phase Extraction/Tandem Mass Spectrometry for Identification of Bioactive Compounds in Mixtures,"
Forsberg, E. M.; Brennan, J. D.,
Analytical Chemistry 86, 8457-8465 (2014). (doi: 10.1021/ac5022166..) - "Continuous Flow Immobilized Enzyme Reactor-Tandem Mass Spectrometry for Screening of AChE Inhibitors in Complex Mixtures.,"
Forsberg, E. M.; Green, J. R. A.; Brennan, J. D.,
Analytical Chemistry 83, 5230-5236 (2011). (doi: 10.1021/ac200534t..)
