Gregory Holland
Professor, Analytical and Physical Chemistry
office: GMCS 213C
phone: 619-594-1596
email: gholland@sdsu.edu

Holland Group Page
Curriculum Vitae
- B.S., Chemistry, State University of New York at Buffalo, 1998
- Ph.D., Analytical and Physical Chemistry, University of Wyoming, 2003
- Postdoctoral Fellow, Sandia National Laboratories, 2003-2006
- Assistant Professor for Research, Arizona State University 2006-2015
- Assistant Professor at San Diego State University 2015-2018
- Associate Professor at San Diego State University 2018-2021
- Professor at San Diego State University 2021-present
Research Interests
The Holland Lab explores the molecular structure and dynamics of complex biological and technologically relevant materials. We are interested in scientific problems that lie at the interface of chemistry, biology and materials science including biologically inspired materials and molecules and nanomaterials functionalized with biomolecules. Although we use a suite of characterization techniques to understand the systems of interest, the primary focus is on developing and applying nuclear magnetic resonance (NMR) methods. A continuing theme in our work is linking the role of molecular structure and dynamical features to material properties and biological function.
 Spider silk:
Spider silk is one of the
toughest materials known. A spider produces this amazing material from an
aqueous spinning dope at ambient temperature and pressure. Spiders produce a
range of different silks with varying mechanical and physical properties. We
have been utilizing a combination of magnetic resonance techniques to
understand the molecular structure of the proteins that comprise the silks and
further, to characterize the silk producing process as a whole. It is our
belief that a better molecular-level understanding of the proteins' structure
and biochemical processing conditions utilized to assemble the silk fiber will
lead to a route for synthetic spider silk production.
Spider silk:
Spider silk is one of the
toughest materials known. A spider produces this amazing material from an
aqueous spinning dope at ambient temperature and pressure. Spiders produce a
range of different silks with varying mechanical and physical properties. We
have been utilizing a combination of magnetic resonance techniques to
understand the molecular structure of the proteins that comprise the silks and
further, to characterize the silk producing process as a whole. It is our
belief that a better molecular-level understanding of the proteins' structure
and biochemical processing conditions utilized to assemble the silk fiber will
lead to a route for synthetic spider silk production.
Lipid Rafts: The viewpoint that cellular membranes are comprised of a continuous liquid crystalline phase of phospholipids is rapidly changing. In the past decade, strong evidence has been presented that supports the presence of heterogeneous domains that are rich in sphingomyelin and cholesterol within the plasma membrane. These domains have been coined "lipid rafts". It has been postulated that these rafts are functional platforms that perform a number of cellular processes. Lipid rafts have been implicated as potential sites for pathogen entry and toxin binding and have been linked to a number of diseases including Alzheimer's, Parkinson's, cardiovascular disease and HIV. We are studying model membrane mixtures that form lipid rafts to understand the molecular interactions involved in phase separation. Recently, we began investigating how protein neurotoxins bind and interact with these complex biological membrane mimics.
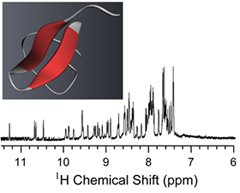 Spider Venom:
Spiders are one of the most successful venomous animals on the planet. There have been over 40,000 species of spider described and each spider's venom contains hundreds to thousands of biologically active components. Our group has been focused on characterizing inhibitor cysteine knot (ICK) neurotoxins extracted from tarantula venom. These ICK peptides bind and modulate a diverse range of ion channels with high selectivity and affinity making them indispensible tools for elucidating the function of ion channels. We are studying the neurotoxin structures with conventional solution NMR and probing how they interact with model lipid membranes and channels with solid-state NMR approaches.
Spider Venom:
Spiders are one of the most successful venomous animals on the planet. There have been over 40,000 species of spider described and each spider's venom contains hundreds to thousands of biologically active components. Our group has been focused on characterizing inhibitor cysteine knot (ICK) neurotoxins extracted from tarantula venom. These ICK peptides bind and modulate a diverse range of ion channels with high selectivity and affinity making them indispensible tools for elucidating the function of ion channels. We are studying the neurotoxin structures with conventional solution NMR and probing how they interact with model lipid membranes and channels with solid-state NMR approaches.
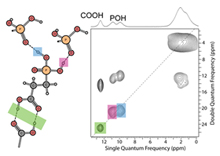 Nanomaterials:
In the past decade, the
synthesis, characterization and application of nanomaterials has become a major
research area in the chemical and biological sciences with numerous research
centers and institutions dedicated to this field. While many synthetic chemists
and materials scientists have produced countless novel nanomaterials, the full
characterization of the products is far less developed. We have been studying
the fundamental chemistries both structural and dynamic at the interface of nanomaterials
with NMR spectroscopy. A combination of solution- and solid-state NMR methods
is implemented to elucidate the capping chemistry and organization of
biomolecular ligands at the surface of nanomaterials. The primary goal of this
work is to develop "easy to implement" NMR techniques to provide
standard structural characterization for functionalized nanoparticles and
nanomaterials.
Nanomaterials:
In the past decade, the
synthesis, characterization and application of nanomaterials has become a major
research area in the chemical and biological sciences with numerous research
centers and institutions dedicated to this field. While many synthetic chemists
and materials scientists have produced countless novel nanomaterials, the full
characterization of the products is far less developed. We have been studying
the fundamental chemistries both structural and dynamic at the interface of nanomaterials
with NMR spectroscopy. A combination of solution- and solid-state NMR methods
is implemented to elucidate the capping chemistry and organization of
biomolecular ligands at the surface of nanomaterials. The primary goal of this
work is to develop "easy to implement" NMR techniques to provide
standard structural characterization for functionalized nanoparticles and
nanomaterials.
Recent Publications
- "Biphasic nature of lipid bilayers assembled on silica nanoparticles and evidence for an interdigitated phase,"
Stengel D., Thai R., Li Y., Peters N.M., Holland G.P.,
Soft Matter 19, 1882-1889 (2023). (doi: 10.1039/d2sm01517j.) - "Tyrosine's Unique Role in the Hierarchical Assembly of Recombinant Spider Silk Proteins: From Spinning Dope to Fibers,"
Stengel D., Saric M., Johnson H.R., Schiller T., Diehl J., Chalek K., Onofrei D., Scheibel T., Holland G.P.,
Biomacromolecules 24, 1463-1474 (2023). (doi: 10.1021/acs.biomac.2c01467.) - "Synthesis and Structural Characterization of Morphologically Distinct Nanoscale Hydroxyapatites,"
Li Y., Deguzman M.A., Heise J.E., Holland G.P.,
Journal of Physical Chemistry C 126, 12790-12801 (2022). (doi: 10.1021/acs.jpcc.2c02383.) - "Assembly and Thermal-Induced Polymerization of Histidine on Fumed Silica Surfaces,"
Swanson H.L., Couri E.A., Sabando C.J.P., Holland G.P.,
ACS Earth and Space Chemistry 6, 1552-1562 (2022). (doi: 10.1021/acsearthspacechem.2c00059.) - "Hydration-Induced β-Sheet Crosslinking of α-Helical-Rich Spider Prey-Wrapping Silk,"
Stengel D., Addison J.B., Onofrei D., Huynh N.U., Youssef G., Holland G.P.,
Advanced Functional Materials 31, 2007161 (2021). (doi: 10.1002/adfm.202007161.) - "The impact of metal doping on fumed silica structure and amino acid thermal condensation catalytic properties,"
Swanson H.L., Pokhrel S., Davidowski S.K., Mädler L., Brinker C.J., Holland G.P.,
Journal of Materials Science 56, 16916-16927 (2021). (doi: 10.1007/s10853-021-06412-0.) - "Investigating the Atomic and Mesoscale Interactions that Facilitate Spider Silk Protein Pre-Assembly,"
Onofrei D., Stengel D., Jia D., Johnson H.R., Trescott S., Soni A., Addison B., Muthukumar M., Holland G.P.,
Biomacromolecules 22, 3377-3385 (2021). (doi: 10.1021/acs.biomac.1c00473.) - "Selective One-Dimensional 13C-13C Spin-Diffusion Solid-State Nuclear Magnetic Resonance Methods to Probe Spatial Arrangements in Biopolymers including Plant Cell Walls, Peptides, and Spider Silk,"
Addison B., Stengel D., Bharadwaj V.S., Happs R.M., Doeppke C., Wang T., Bomble Y.J., Holland G.P., Harman-Ware A.E.,
Journal of Physical Chemistry B 124, 9870-9883 (2020). (doi: 10.1021/acs.jpcb.0c07759.) - "Probing the binding modes and dynamics of histidine on fumed silica surfaces by solid-state NMR,"
Swanson H.L., Guo C., Cao M., Addison J.B., Holland G.P.,
Physical Chemistry Chemical Physics 22, 20349-20361 (2020). (doi: 10.1039/d0cp03472j.) - "Fusion of Bipolar Tetraether Lipid Membranes Without Enhanced Leakage of Small Molecules,"
Geoffray Leriche, Dillan Stengel, David Onofrei, Takaoki Koyanagi, Gregory P. Holland, and Jerry Yang,
Scientific Reports 9, 19359 (2019). (10.1038/s41598-019-55494-z.) - "H-2 NMR reveals liquid state-like dynamics of arene guests inside hexameric pyrogallol[4]arene capsules in the solid state,"
Irazema J. Islas, Dillan Stengel, Cesar A. Garcia, J. Bennett Addison, George N. Samaan, Gregory P. Holland, and Byron W. Purse,
Org. Chem. Front. 6, 1361-1366 (2019). (doi: 10.1039/C9QO00232D.) - "Entropic effects enable life at extreme temperatures,"
Young Hun Kim, Geoffray Leriche, Karthik Diraviyam, Takaoki Koyanagi, Kaifu Gao, David Onofrei, Joseph Patterson, Anirvan Guha, Nathan Gianneschi, Gregory P. Holland, Michael K. Gilson, Michael Mayer, David Sept, Jerry Yang,
Sci. Advances 5, eaaw4783 (2019). (DOI: 10.1126/sciadv.aaw4783.) - "Experimental Methods for Characterizing the Secondary Structure and Thermal Properties of Silk Proteins,"
Meghan McGill, Gregory P. Holland, and David L. Kaplan,
Macromolec. Rapid Comm. 40, 1800390 (2019). (doi: 10.1002/marc.201800390.) - "Investigating the Interaction of Grammostola rosea Venom Peptides and Model Lipid Bilayers with Solid-state NMR and Electron MIcroscopy Techniques,"
G. Polido, X. Shi, D. Xu, C. Guo, R. Thai, J.P. Patterson, N.C. Gianneschi, T.M. Suchyna, F. Sachs, G.P. Holland,
Biochim. Biophys. Acta 1861, 151-160 (2019). (doi: 10.1016/j.bbamem.2018.08.004.)
