Rees Garmann
Assistant Professor, Physical Chemistry, Biochemistry, Biophysics, Viromics
office: CSL 334
email: rgarmann@sdsu.edu

Garmann Group Page
Curriculum Vitae
- B.Sc., Chemistry, U.C. Berkeley, 2007
- Ph.D., Physical Chemistry, UCLA, 2014
- Postdoc, Harvard University, 2015-2020
- Assistant Professor, San Diego State University, 2021-
Research Interests
Our research group studies how viruses—the baddest parasites on the planet—use molecular interactions to infect their hosts, replicate, and evolve. Our approach is different from that of most other virology groups in that we often work with viruses outside of the host cell, where we can use physical chemistry techniques to zoom in on the virus particles and make very sensitive measurements of their essential properties.
We take viruses apart, we separate their protein and nucleic acid subunits, and then we put them back together. We measure the structure and dynamics of the subunits in isolation, and we also measure collective properties that emerge when many subunits interact. We team up with theorists and computational chemists to turn our measurements into quantitative models that help us understand how viruses perform complex biological functions, such as assembling, uncoating, and regulating gene expression.
In addition to teaching us about how viruses work, our results can inspire new ways of repurposing viruses to do useful things, such as building nanoscale materials or delivering therapeutic cargos to cells. And most importantly, our results can also inform new ways to block the spread of harmful viruses that cause disease.
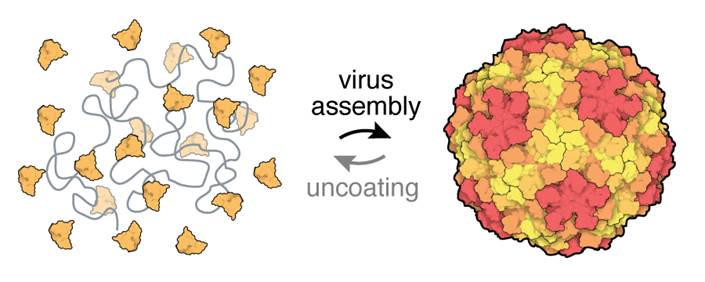
Because our approach borrows from many fields of basic and applied science—including molecular biology, chemistry, physics, and nanoscience—students in the group will necessarily be exposed to a broad range of experimental and theoretical techniques. These techniques include traditional molecular biology, such as cloning, protein expression, and nucleic acid synthesis; biophysical techniques, such as electrophoresis, sedimentation, and spectroscopy; structural biology methods, such as electron microscopy and RNA structure prediction; methods from nanoscience, including surface chemistry and nanoparticle functionalization; methods from DNA nanotechnology, such as DNA origami and strand exchange reactions; and nanoscale optical imaging techniques, in particular interferometric scattering microscopy. In addition, when established techniques fail, we're not afraid to invent new techniques ourselves.
If any of this stuff sounds interesting, don't hesitate to email Prof. Rees Garmann at rgarmann@sdsu.edu.
Selected Publications
- "A simple RNA-DNA scaffold templates the assembly of monofunctional virus-like particles,"
Garmann, R.F., Sportsman, R., Beren, C., Manoharan, V.N., Knobler, C.M. and Gelbart, W.M.,
Journal of the American Chemical Society 137(24), 7584-7587 (2015). (doi: 10.1021/jacs.5b03770.) - "Physical principles in the self-assembly of a simple spherical virus,"
Garmann, R.F., Comas-Garcia, M., Knobler, C.M. and Gelbart, W.M.,
Accounts of chemical research 49(1), 48-55 (2016). (doi: 10.1021/acs.accounts.5b00350.) - "Dynamic measurements of the position, orientation, and DNA content of individual unlabeled bacteriophages,"
Goldfain, A.M., Garmann, R.F., Jin, Y., Lahini, Y. and Manoharan, V.N.,
Journal of Physical Chemistry B 120(26), 6130-6138 (2016). (doi: 10.1021/acs.jpcb.6b02153.) - "Measurements of the self-assembly kinetics of individual viral capsids around their RNA genome,"
Garmann, R.F., Goldfain, A.M. and Manoharan, V.N.,
Proceedings of the National Academy of Sciences 116(45), 22485-22490 (2019). (doi: 10.1073/pnas.1909223116.) - "Single-particle studies of the effects of RNA-protein interactions on the self-assembly of RNA virus particles,"
Garmann R.F., Goldfain A.M., Tanimoto C.R., Beren C.E., Vasquez F.F., Villarreal D.A., Knobler C.M., Gelbart W.M., Manoharan V.N.,
Proc. Natl. Acad. Sci. 119, e2206292119 (2022). (doi: 10.1073/pnas.2206292119.) - "Multiple Nucleocapsid Structural Forms of Shrimp White Spot Syndrome Virus Suggests a Novel Viral Morphogenetic Pathway,"
Huang H.-J., Tang S.-L., Chang Y.-C., Wang H.-C., Ng T.H., Garmann R.F., Chen Y.-W., Huang J.-Y., Kumar R., Chang S.-H., Wu S.-R., Chao C.-Y., Matoba K., Kenji I., Gelbart W.M., Ko T.-P., Wang H.-J., Lo C.-F., Chen L.-L., Wang H.-C,
Intl. J. Molec. Sci. 24, 7525 (2023). (DOI: 10.3390/ijms24087525.)
