
Love Group Page
Research Interests
The overall focus of the research conducted in my lab is protein design and engineering. Our initial engineering goal is to redesign, mutate and drive small proteins to self-assemble into complexes of specific structure (e.g. precise dimer formation). The small proteins will then be redesigned to bind to specific regions of target proteins expressed by pathogenic organisms. These design targets are geared towards applications in the field of protein-based drug design. Our approach entails a design cycle which couples computer science, molecular biology, biochemistry and spectroscopy. This cyclic approach ultimately increases our understanding of the forces that drive protein self-assembly and raises the likelihood of successful protein-protein complex design.
Computational Approach: The first step in driving de novo protein self-assembly is the computational docking of the two molecules in a predefined orientation (see figure on left). The algorithms we have developed utilize an established geometric recognition algorithm that treats the molecules as rigid bodies and rigorously assesses interfacial surface complementarity as a function of translational and rotational position. This process is computationally intensive yet has been rendered tractable upon incorporation of the Fourier correlation theorem. Once we obtain the optimal intermolecular atomic coordinates, the two molecules are treated as one and a suite of highly-developed protein-design algorithms (collectively termed ORBIT) is used to repack the interfacial side-chains in a manner analogous to the cores of well-folded proteins. The ORBIT algorithms, which utilize advanced molecular-mechanics force fields, return a mutated amino-acid sequence that has been optimized to drive precise complex formation through favorable hydrophobic, hydrophilic and electrostatic interactions.
Molecular Biology: Tools of molecular biology (e.g. recursive PCR, T7 bacterial expression) are used to introduce the mutations and physically generate the redesigned proteins. We are also planning to utilize a genetic screen/combinatorial process (i.e., a bacterial two-hybrid screen) to quickly assess and enhance the success of docking.
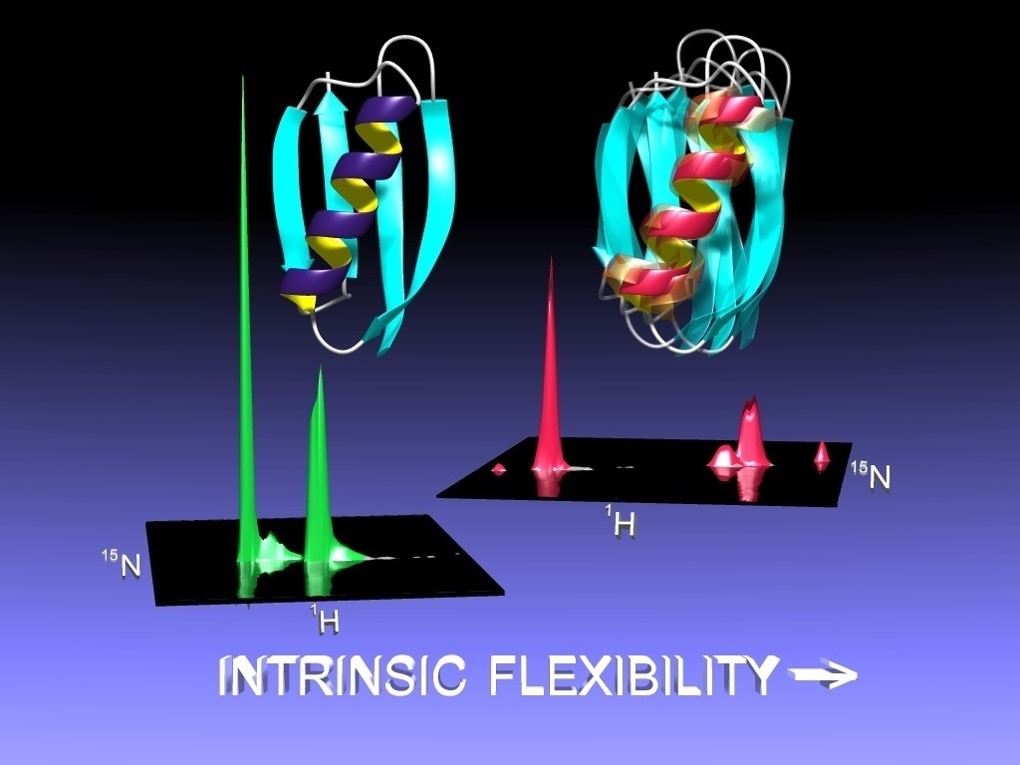
Biochemistry and Spectroscopy: Various biochemical methods and techniques are used to purify the expressed proteins (e.g. FPLC and HPLC) and further assess the degree of complex formation (e.g. size-exclusion chromatography, analytical ultracentrifugation). Finally, to verify that the designed proteins are docked in the target orientation, the structure of each complex is solved by either multidimensional NMR or x-ray crystallography.
Selected Publications
- "Effect of Ultraviolet Light Exposure and Compost Tea Supplementation on Growth, Antioxidant Activities, and Microbiome of Hydroponically Grown Mustard Greens,"
Maniaci B., Lipper C.H., Anipindi D.L., Erlandsen H., Cole J.L., Stec B., Huxford T., Love J.J,
Biochem 58, 2199-2207 (2019). (10.1021/acs.biochem.9b00055.) - "Kayla J Hurd, Shruti Shertukde, Trevor Toia, Angelina Trujillo, Ramona L Pérez, David L Larom, John J Love, Changqi Li,"
The Cultural Importance of Edible Insects in Oaxaca, Mexico,
Annals Entomol. Soc. Am 112, 552-559 (2019). (doi: 10.1093/aesa/saz018.) - "Romo-Fewell, Octavio; Navarro, Mario; Love, John J,"
Ubiquitin Carboxy Hydrolase-L3 (UCH-L3): A Molecular Sensor of Protein Stability?,
FASEB J 24 (2010).
